New assay assesses multiple cellular pathways at once

A novel technological approach developed by researchers at Baylor College of Medicine expands from two to six the number of molecular pathways that can be studied simultaneously in a cell sample with the dual luciferase assay, a type of testing method commonly used across biomedical fields.
Published in the journal Nature Communications, the report shows that multiplexed hextuple luciferase assaying, meaning a testing method that can effectively probe six different pathways. It can also be used to monitor the effects of experimental treatments on multiple molecular targets acting within these pathways. The new assay is sensitive, saves time and expense when compared to traditional approaches, reduces experimental error and can be adapted to any research field where the dual luciferase assay is already implemented, and beyond.
“One of the interests of our lab is to have a better understanding of the processes involved in cancer. Cancer usually originates through changes on many different genes and pathways, not just one, and currently most cell-based screening assays conduct single measurements,” said corresponding author Dr. Koen Venken, assistant professor of biochemistry and molecular biology, and pharmacology and chemical biology at Baylor.
To get a more detailed picture of the cellular processes that differentiate normal versus cancer cells, researchers resort to conduct several independent screening assays at the expense of time and additional cost.
“Our goal in this study was to measure multiple cellular pathways at once in a single biological sample, which would also minimize experimental errors resulting from conducting multiple separate assays using different samples,” said Venken, a McNair Scholar and member of the Dan L Duncan Comprehensive Cancer Center at Baylor.
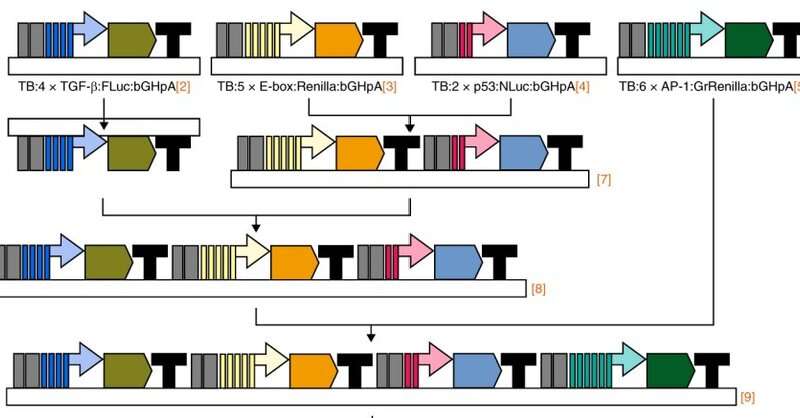
Dr. Alejandro Sarrion-Perdigones, first author of the paper, focused on developing a multiplexed method—a method for simultaneously detecting many signals from complex systems, such as living cells. He developed a sensitive assay using luciferases, enzymes that produce bioluminescence. The assay includes six luciferases, each one emitting bioluminescence that can be distinguished from the others. Each luciferase was engineered to reveal the activity of a particular pathway by emitting bioluminescence.
“To engineer and deliver the luciferase system to cells, we used a ‘molecular Lego’ approach,” said co-author Dr. Lyra Chang, post-doctoral researchers at the Center for Drug Discovery at Baylor. “This consists of connecting the DNA fragments encoding all the biological and technological information necessary to express each luciferase gene together sequentially forming a single DNA chain called vector. This single vector enters the cells where each luciferase enzyme is produced separately.”
Treating the cells with a single multi-luciferase gene vector instead of using six individual vectors, decreased variability between biological replicates and provided an additional level of experimental control, Chang explained. This approach allowed for simultaneous readout of the activity of five different pathways, compared to just one using traditional approaches, providing a much deeper understanding of cellular pathways of interest.
Source: Read Full Article